Research
The Role of the Tumor Microenvironment in Ovarian Cancer
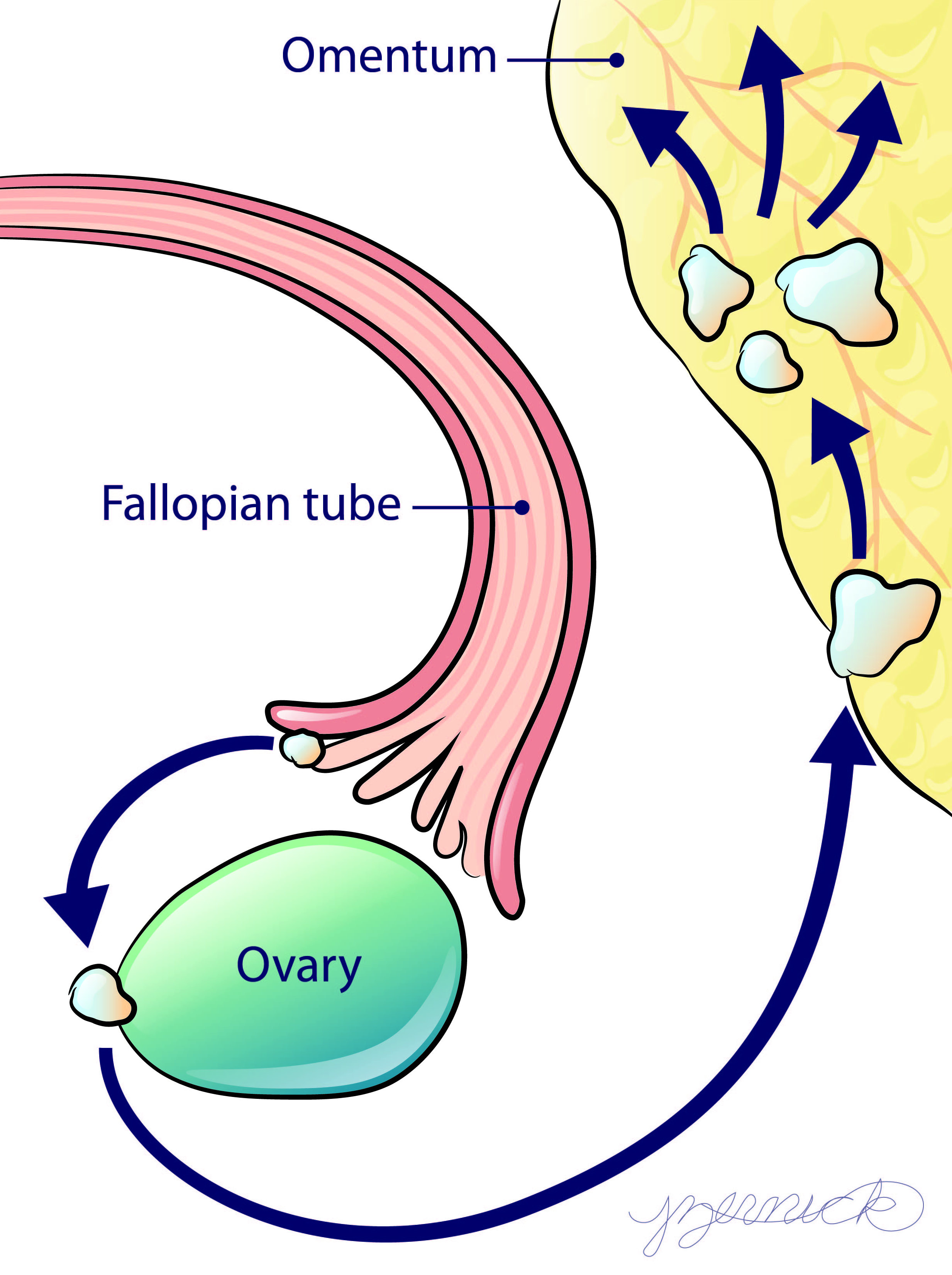
High-grade serous ovarian cancer (HGSOC) commonly inititates on the ends of the fallopian tube before it progresses to the ovary and peritoneal tissues such as the omentum. During this progression the tumor cells interact with diverse microenvironments that may influence the ability for the tumor to grow and respond to treatment. Our lab has engineered several in vitro biomimetic systems to understand the influence of these diverse microenvironments. From studies in these systems, we seek to identify predictors of tumor sensitivity to existing therapies as well as new therapeutic targets to improve patient palliation and/or outcome.
Image courtesy Jennifer Zernick.
Theme 1: How does the extracellular matrix (ECM) influence HGSOC progression?

Collaborators: Kristyn Masters, Alexandra Naba, Suzanne Ponik
Theme 2: How do tumor cells disseminate during metastasis?

Collaborators: Lisa Baroillhet, Stephanie McGregor, Alejandro Roldan-Azate
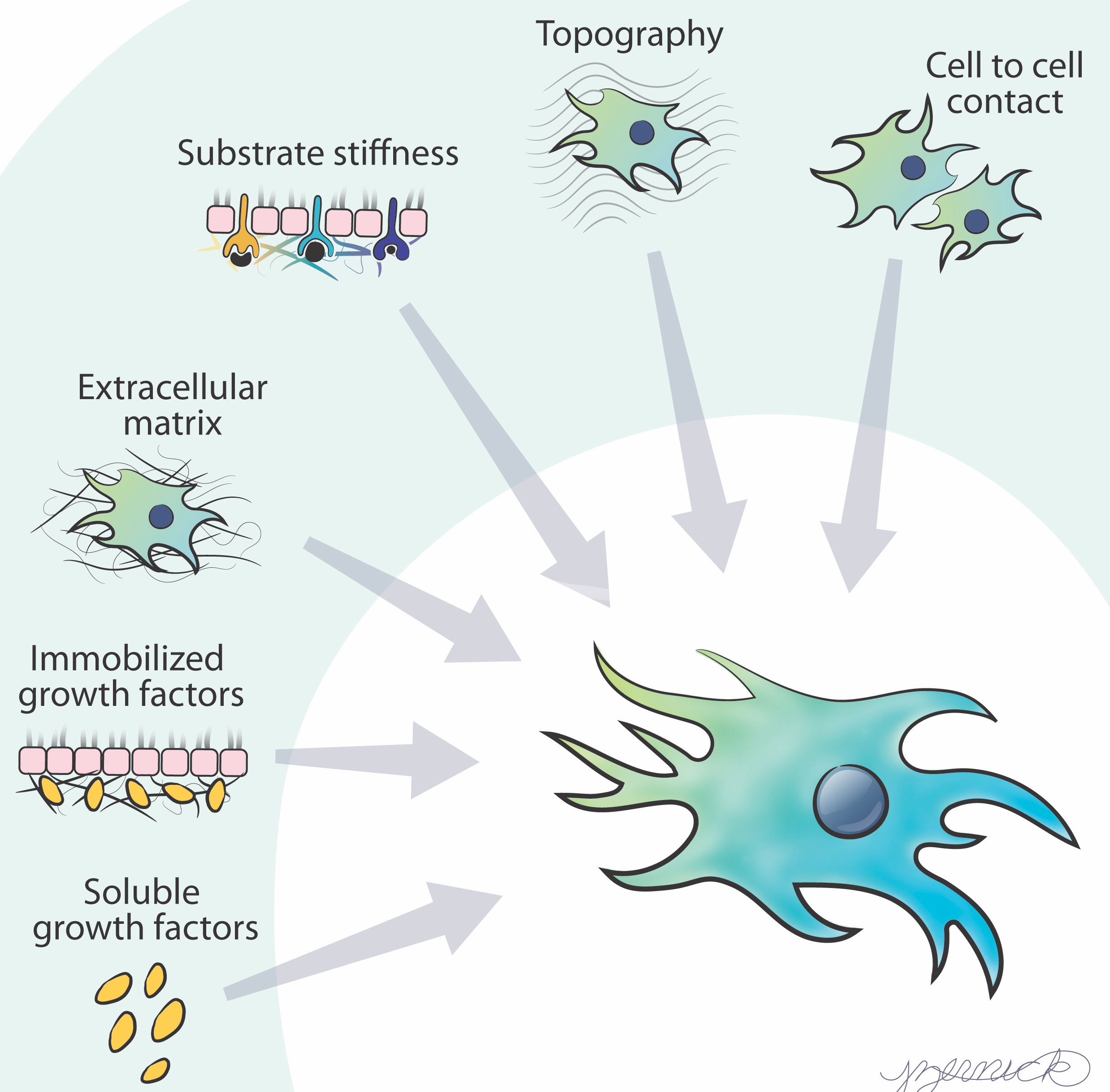
Cellular Decision-Making
Image courtesy Jennifer Zernick.
Theme 1: How do cells integrate information to direct collective migration?
Collaborators: Kristyn Masters, Jacob Notbohm.
Theme 2: How do cells interpret changes in growth factor presentation?
Collaborators: Megan McClean, Kristyn Masters.